1 min read
Antibodies Targeting Three Spike Protein Domains Regulating SARS-CoV-2 Infectivity
By Antibody Solutions Research Team on May 11, 2021 1:57:00 PM
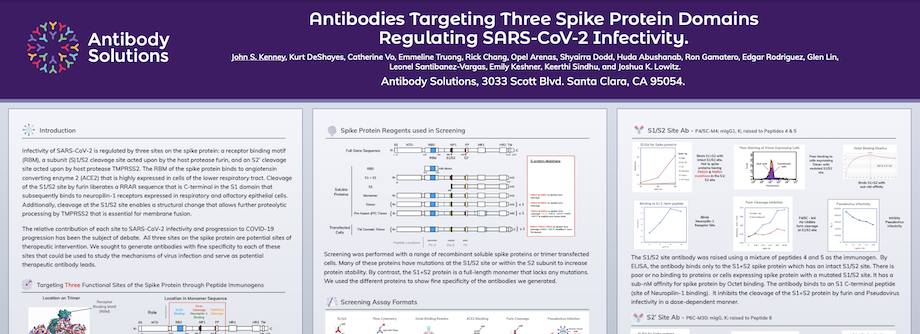
Infectivity of SARS-CoV-2 is regulated by three sites on the spike protein: a receptor binding motif (RBM), a subunit (S)1/S2 cleavage site acted upon by the host protease furin, and an S2’ cleavage site acted upon by host protease TMPRSS2. The RBM of the spike protein binds to angiotensin converting enzyme 2 (ACE2) that is highly expressed in cells of the lower respiratory tract. Cleavage of the S1/S2 site by furin liberates a RRAR sequence that is C-terminal in the S1 domain that subsequently binds to neuropilin-1 receptors expressed in respiratory and olfactory epithelial cells. Additionally, cleavage at the S1/S2 site enables a structural change that allows further proteolytic processing by TMPRSS2 that is essential for membrane fusion.
Topics: SARS-CoV-2 Posters
1 min read
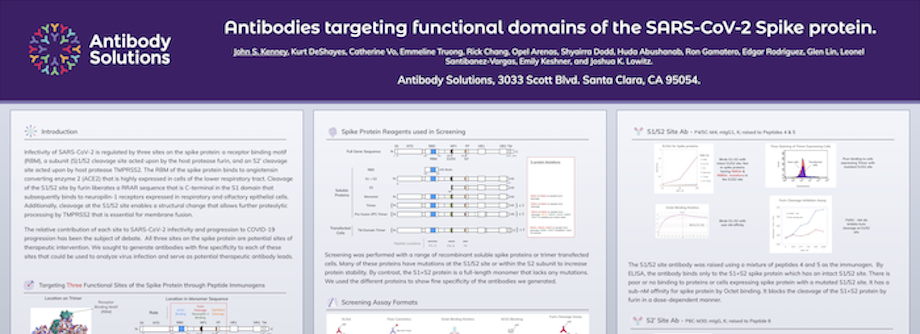
Antibody targeting functional domains of the SARS-CoV-2 Spike protein
By Antibody Solutions Research Team on Jan 4, 2021 10:17:00 AM
Infectivity of SARS-CoV-2 is regulated by three sites on the spike protein: a receptor binding motif (RBM), a subunit (S)1/S2 cleavage site acted upon by the host protease furin, and an S2’ cleavage site acted upon by host protease TMPRSS2. The RBM of the spike protein binds to angiotensin converting enzyme 2 (ACE2) that is highly expressed in cells of the lower respiratory tract. Cleavage of the S1/S2 site by furin liberates a RRAR sequence that is C-terminal in the S1 domain that subsequently binds to neuropilin-1 receptors expressed in respiratory and olfactory epithelial cells. Additionally, cleavage at the S1/S2 site enables a structural change that allows further proteolytic processing by TMPRSS2 that is essential for membrane fusion.
Topics: SARS-CoV-2 Posters
Filter by Keyword
- Posters (21)
- Publications (15)
- Therapeutic Monoclonal Antibodies (4)
- Monoclonal (3)
- Multi-meric Membrane (3)
- Multi-pass transmembrane (3)
- Transgenic Animals (3)
- ELISAs (2)
- Human Therapeutic Antibodies (2)
- Hybridoma (2)
- SARS-CoV-2 (2)
- APOBEC3G (1)
- ARMER (1)
- ATX-GX (1)
- Alloy Therapeutics (1)
- Antibody Discovery (1)
- Antibody Generation (1)
- BRCA2 (1)
- CEM15 (1)
- Cadherin-11 (1)
- Carcinogenesis (1)
- Conditioned Media (1)
- Critical Reagents (1)
- Cystine Knot Peptides (1)
- D Protein (1)
- DLL4 (1)
- Fully Human (1)
- HIV-1 (1)
- HSA (1)
- IL-1 alpha (1)
- Immune B-cells (1)
- Immunization (1)
- Knottins (1)
- L-amino acids (1)
- L-selectin (1)
- LAM (1)
- LBAs (1)
- LOXL2 (1)
- Lymphangioleiomyomatosis (1)
- McAbs (1)
- OmniAb (1)
- OmniRat (1)
- PNAd (1)
- Pharmacokinetic (PK) (1)
- Prolactin (1)
- Secretion Capture Report Web (1)
- Stereochemistry (1)
- Therapeutic Targets (1)
- Tissue Culture (1)
- Transgenic H2L2 Mice (1)
- VEGF-A (1)
- VEGF-C (1)
- VEGF-D (1)
- p53 (1)