2 min read
The UK Strain and Other Recent RBD Mutations in the SARS-CoV-2 Virus
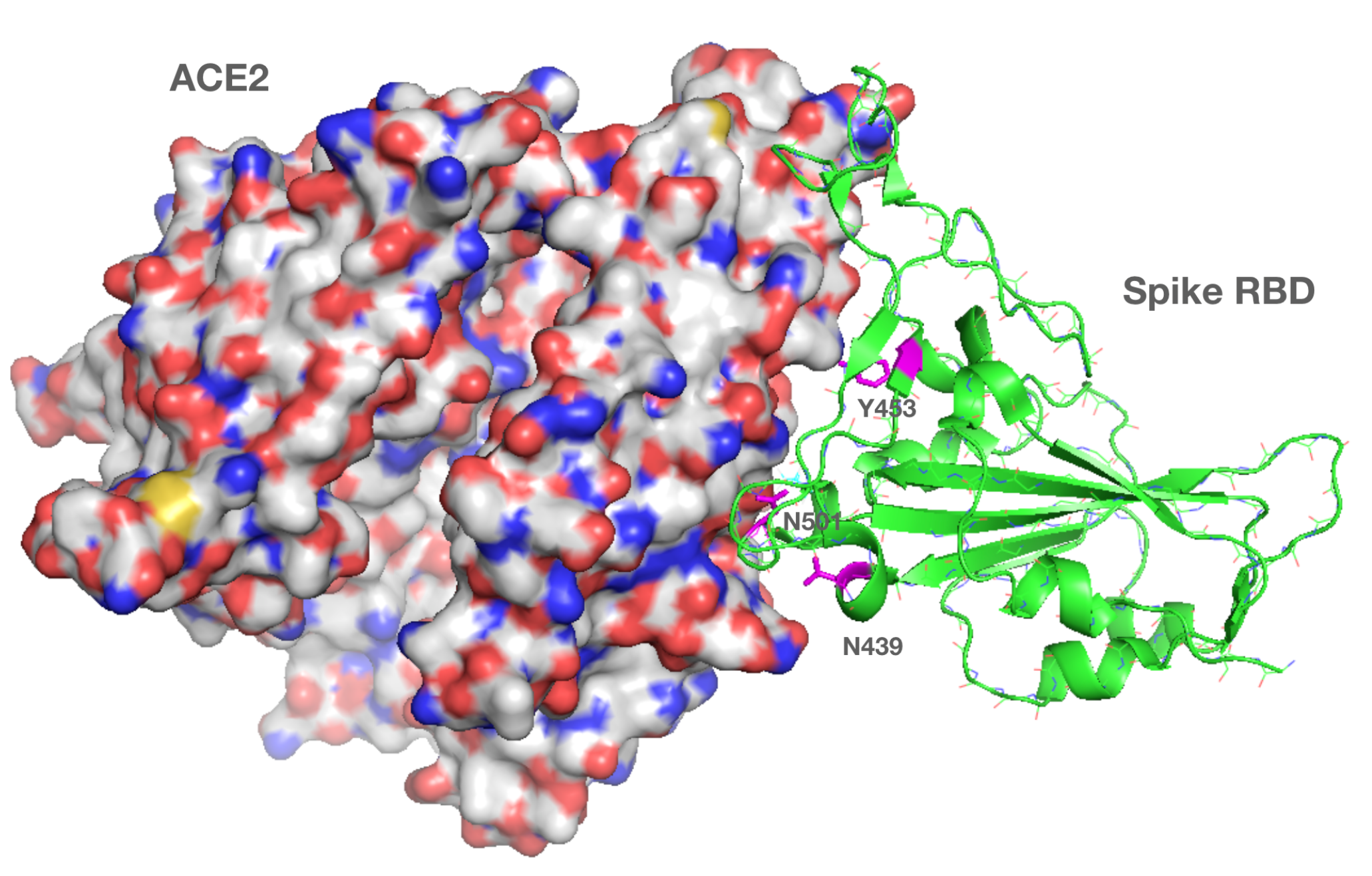
Examining mutations in the SARS-CoV-2 Spike (S) Protein Receptor Binding Domain (RBD) at the ACE2 receptor S protein interface
We’ve reported on the SARS-CoV-2 S protein mutant D614G in past blog posts and the potential impact of the mutation on infectivity and furin processing. While mutations occur commonly in RNA viruses, most do not provide an advantage to the virus. Recently, scientists in the United Kingdom have described a lineage of the coronavirus, designated B.1.1.7, that appears to be more transmissible than other more established lineages. The B.1.1.7 lineage has a mutation in the receptor binding domain (RBD) of the Spike protein at position 501, where amino acid asparagine (N) has been replaced with tyrosine (Y). Several other mutations in the S protein have garnered attention. A N439K mutation, that first emerged in Scotland in lineage B.1.141, is now circulating in many European countries and internationally. The Y453F mutation has been observed in the context of mink-human infections, and it appeared in the widely reported Danish Mink cluster.
Using our in-house modeling software, we have identified the sites of the three S protein mutations (e.g., N501, N439 and Y453) in the structure reported for the S protein bound to the ACE2 receptor.1 It is noteworthy that the sites of all three mutations are observed to lie at the interface between the ACE2 and RBD. Each mutation, alone or in concert, may act to increase the affinity of the ACE2:RBD interaction, a key step in virus infection. Specifically, N501Y has been reported to increase the binding affinity of S protein to human and murine ACE2. Additionally, the mutations may change the way the virus interacts with the immune system or the response generated by a vaccine.
Location of three S protein mutations (shown as magenta) at the ACE2 receptor S protein interface found in SARS-CoV-2 variants
Having identified the sites of mutation within the S protein RBD, we will be using modeling and docking software to evaluate the mutations and how they impact the molecular interaction between the S protein and the ACE2 cell surface receptor.
It is important to point out that, at this time, there is no evidence that these mutations cause more severe illness, increased risk of death, or affect vaccine efficacy. However, with a mutation rate estimated to be of about one to two changes per month, the evolving nature of the SARS-CoV-2 virus may require new diagnostic, vaccine and therapeutic developments for years to come.
Reference(s):
1Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., Wang, X. (2020) Nature 581: 215-220.

Author of more than 40 publications, John’s current research interests include new technologies for improving therapeutic antibody discovery, properties of next-generation antibody-like molecules, and best practices for critical reagents used in biologics development.