6 min read
COVID-19 Targets, Tools and Therapeutics (Part 2)
Antibody Solutions is doing its part to respond to the COVID-19 pandemic, including original research, tool discovery, and participation in in-house and client-sponsored discovery programs. In this new blog series, we examine questions such as: What are the potential targets to treat COVID-19 and its sequelae, and what are the strategies and tools to discover drugs to treat the disease?(In Part 1 of this blog series, we outlined the causes of COVID-19 and disease targets. In Part 2, we describe the tools that can be used to study the targets and discover potential therapies. In Part 3, coming soon, we’ll discuss the potential therapies that are currently under consideration.)
The multitude of patient responses to SARS-CoV-2 infection, manifestations of COVID-19, and its sequelae will require a diverse set of interventions targeting the virus, the host response to the virus, and the organ-specific medical conditions that arise. In this installment we describe the tools, methods and systems needed to validate the targets of COVID 19 and its sequelae, and to further identify potential vaccines and therapies.
COVID-19 Reagents
The rapid availability of a diverse portfolio of research reagents is proving essential for accelerating Covid-19 scientific discovery. These include commercially available proteins (e.g., spike proteins, soluble receptors and proteases), antibodies targeting the proteins, cell lines expressing viral proteins and receptors, and SARS-CoV-2 pseudovirus. Commercially available research reagents are generally of good quality, but should be vetted for the particular application. In our own investigations, differences have been observed in the binding activity of commercial SARS-CoV-2 reagents depending upon the reagents used and the assay format. This may be a reflection of the epitopes available on the different proteins either occurring naturally or resulting from recombinant expression.
Both SARS‐CoV‐1 and SARS‐CoV‐2 induce highly pathogenic infectious diseases, which need to be operated in a biosafety level 3 (BSL‐3) laboratory for biosafety consideration. Non-pathogenic pseudoviral particles expressing the SARS‐CoV spike protein have similar infection characteristics to the virus but no amplification ability. Using pseudovirus enables studies of infectivity and the rapid assessment of neutralizing antibodies and other proteins in a BSL‐2 laboratory.
Commercial reagents have broad utility for studying what mechanisms are critical for the virus to infect host cells and identify inhibitors of virus infection. However, it is important to validate the functional activity of the reagents in each assay type, method and system of study.
Tools to study SARS-CoV-2 Infectivity
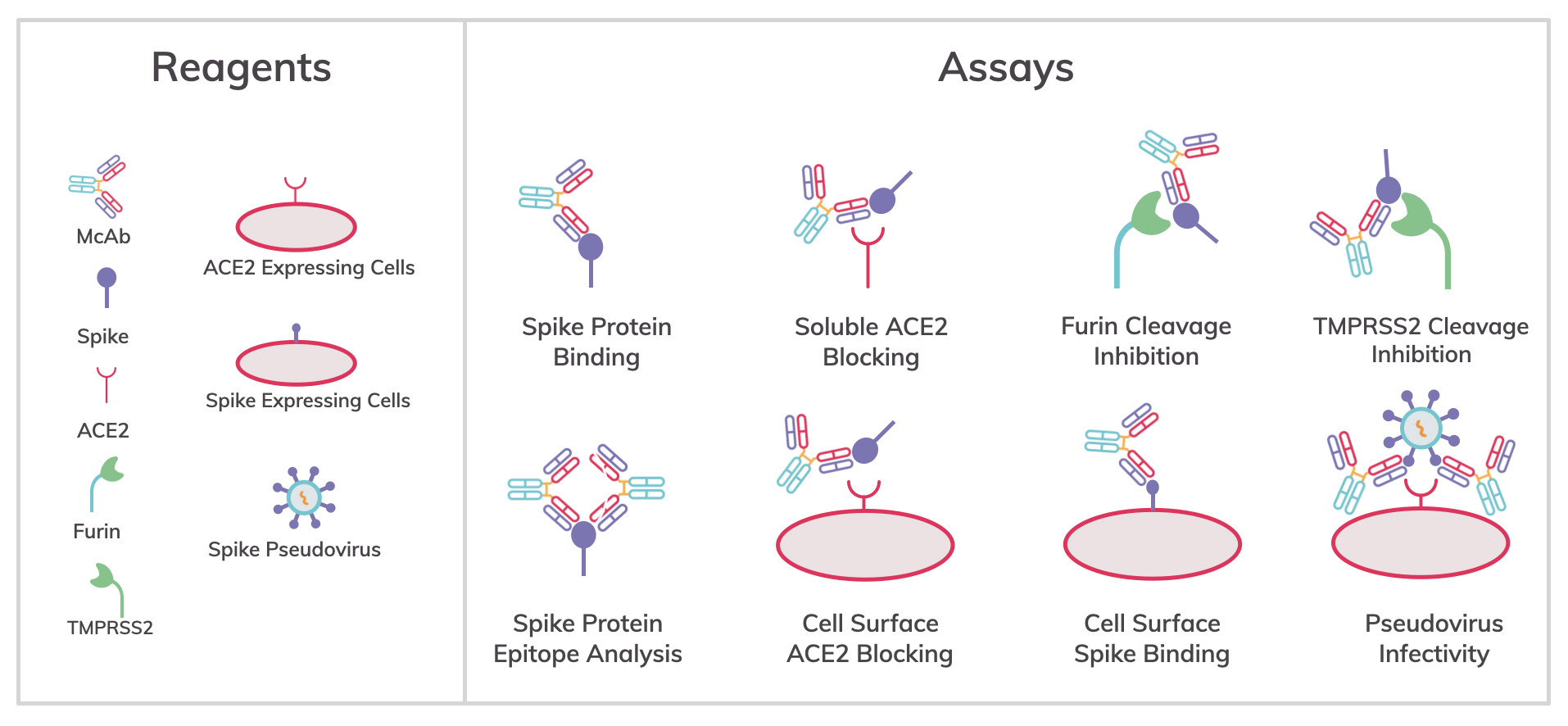
Analysis of mechanisms and inhibitors of SARS-CoV-2 infection
Protein- and cell-based assays are commonly used to analyze one of the three domains of the spike protein (i.e., receptor binding [RBD], and furin, and TMPRSS cleavage) involved in infectivity, and are screening tools and methods to find inhibitors of a specific mechanism:
Ligand-binding assays such as ELISA can identify antibodies binding to the spike protein and biomolecular interaction analysis (BMIA) using.
-
Biacore and Octet instruments can provide affinity measurements and epitope binning of antibodies.
-
Assays using either soluble angiotensin-converting enzyme 2 (ACE2) or cells expressing ACE2 can identify antibodies that block spike RBD domain binding to ACE2.
-
Not to be overlooked, host cell proteases furin and TMPRSS2 arm the virus for fusion with host cell membranes.
-
SDS-Gels and Western blots have been used to understand the proteolytic processing of the spike protein.
-
Assays for enzymatic cleavage of the spike protein can be used to screen antibodies that bind to spike cleavage domains and protect the epitope from cleavage or screen direct active site inhibitors of furin and TMPRSS2 proteolytic activity.
-
Cell surface-expressed spike protein may be used in cell-based flow cytometry assays to identify antibodies binding spike oligomers on host cells.
-
ACE2, furin and TMPRSS2-expressing cells can be used in flow cytometry assays to detect antibodies or other agents that block infection by SARS-Cov-2 pseudovirus particles.
-
Finally, sequencing of viral genomes has been critical in identifying mutations in SARS-CoV-2 likely to impact infectivity.
System & Methods to Study COVID-19 Diseases and Therapies
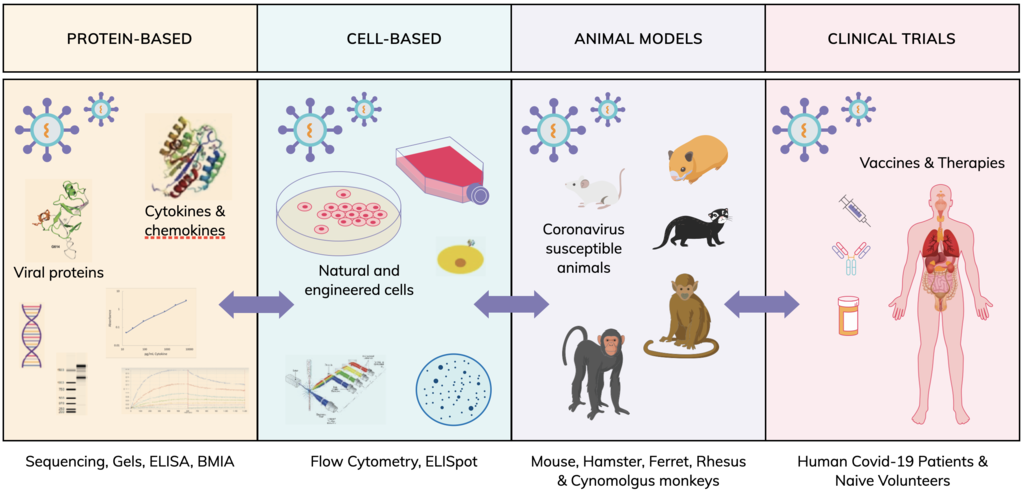
Animal models (in vivo veritas)
Transgenic mice, designated K18-hACE2, have been developed that express human (h)ACE2, in the epithelia of tissues and organs, including lungs, liver, kidney, spleen, heart and gut. They are highly susceptible to SARS-CoV infection and experience high viral lung titres, significant weight loss and morbidity. Mouse-adapted SARS-CoV-2 has been reported in a preprint, with mutations in the receptor binding domain (RBD) of the spike protein following serial passage, inducing productive infection of both young and aged WT BALB/c mice. Both humans and mice display similar clinical signs such as weight loss and pneumonia. Severe infections are often associated with increased pro-inflammatory cytokine production, accompanied by high viral lung titers which correlates with extensive lung damage and significant pulmonary decline. Hamsters, ferrets and non-human primates have also been identified as models to study both SARS-CoV-2 infection and progression of COVID-19 disease and sequelae.
Analysis of the immune response to SARS-CoV-2
ELISAs for viral proteins can be used to measure the humoral immune response to viral vaccines. For COVID-19, SARS-CoV-2 replication in the host can result in the creation of a cytokine storm, pro-inflammatory cytokines such as TNF-α, IFN α / β, IFNγ, IL-1β IL-6, IFNγ, CXCL10 and CCL2. ELISA can be used to assess these cytokine responses to infection and modulation by therapies.
Early work on COVID-19 patients indicates lymphopenia with a substantial reduction in CD4+ T cells, CD8+ T cells and total T cells. Others have reported that functional exhaustion of T cells and NK cells at the earlier stages of SARS-CoV-2 infection may contribute to a reduced ability to eliminate the virus. Flow cytometric analysis is a key tool for characterization of immune cell subsets and their activation states. The ELISpot (enzyme-linked immunospot) assay can be used to measure the number of antibody-secreting from B- cells. Similarly, the T cell response can also be assessed with ELISpot, by measuring the number of cytokine-secreting cells.
Analysis of sequelae
COVID-19 may lead to other medical conditions as a consequence of the initial disease (e.g., sequelae), which can be exacerbated by another disorder (e.g., comorbidities); the specialized reagents and assays required to analyze the various sequelae are too numerous to describe here. However, similar types of tools, systems and methods can be employed to understand the origins of the sequelae and identify therapeutic interventions.
Clinical trials
Ultimately, clinical trials of vaccines and therapies will provide the best understanding of how COVID-19 may be prevented or treated.
But it’s the knowledge gained through protein- and cell-based systems as well as animal models that enable clinical trials to occur. In a similar way, serum, cell and tissue samples generated in a clinical setting may be analyzed in protein- and cell-based systems and in animal models to give a deeper understanding of the mechanisms of SARS-CoV-2 infectivity and COVID-19 progression.
If your team is working on COVID-19 or any of its sequelae and would benefit from having an antibody team at your side, please reach out to us. Tools we’re putting to work in SARS-CoV-2 research include:
We’d love to hear about the directions your studies are taking and what findings are piquing your curiosity. It’s clear that sharing insights among the scientific community can and will help defeat this pathogen.

Author of more than 40 publications, John’s current research interests include new technologies for improving therapeutic antibody discovery, properties of next-generation antibody-like molecules, and best practices for critical reagents used in biologics development.




